【R】Tidymodelsで機械学習 2
2020年5月25日
1.はじめに
こちらも、正直、良くわかっていないのですが、Julia SilgeさんのYoutubeの解説”Get started with tidymodels using vaccination rate data”を見ていたら、わかったような気になったので、とりあえず書いておきます。
RのパッケージTidyverseはデータハンドリングや可視化のツールとして認知されていますが、それと親和性が良い(?)Tidymodelsというパッケージがあります。今回もこれを使います。
2.データ
アメリカの学校のMMR(新三種混合ワクチン. はしか(Measles)、おたふくかぜ(Mumps)、風しん(Rubella)の3疾病を一度に予防する混合ワクチン)の摂取状況のデータです。アメリカ各州のワクチン接種率を予測します。閾値は95%らしいです。
# vaccination rate data
library(tidyverse)
measles <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-02-25/measles.csv')
measles_df <- measles %>%
filter(mmr >0 ) %>%
transmute(state,
mmr_threshold = case_when(mmr > 95 ~ "Above",
TRUE ~ "Below")) %>%
mutate_if(is.character, factor)
library(skimr)
skim(measles_df)
> skim(measles_df)
-- Data Summary ------------------------
Values
Name measles_df
Number of rows 44157
Number of columns 2
_______________________
Column type frequency:
factor 2
________________________
Group variables None
-- Variable type: factor ---------------------------------------------------------------------------------------------
# A tibble: 2 x 6
skim_variable n_missing complete_rate ordered n_unique top_counts
* <chr> <int> <dbl> <lgl> <int> <chr>
1 state 0 1 FALSE 21 Cal: 14225, Ill: 7686, New: 4159, Ohi: 2919
2 mmr_threshold 0 1 FALSE 2 Abo: 31007, Bel: 13150
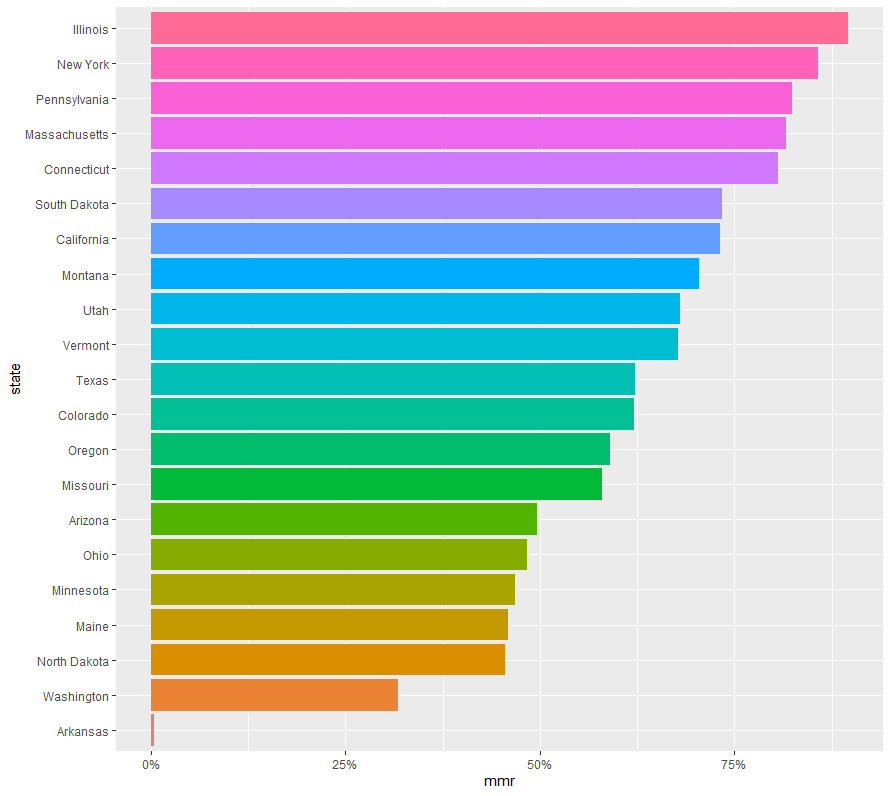
> measles_df %>% group_by(state) %>% summarise(mmr = mean(mmr_threshold == "Above")) %>% arrange(-mmr)
# A tibble: 21 x 2
state mmr
<fct> <dbl>
1 Illinois 0.896
2 New York 0.858
3 Pennsylvania 0.824
4 Massachusetts 0.817
5 Connecticut 0.806
6 South Dakota 0.735
7 California 0.731
8 Montana 0.705
9 Utah 0.680
10 Vermont 0.678
# ... with 11 more rowsmeasles_df %>% group_by(state) %>% summarise(mmr = mean(mmr_threshold == "Above")) %>% arrange(-mmr)
# A tibble: 21 x 2
state mmr
<fct> <dbl>
1 Arkansas 0.00353
2 Washington 0.318
3 North Dakota 0.456
4 Maine 0.459
5 Minnesota 0.468
6 Ohio 0.484
7 Arizona 0.496
8 Missouri 0.580
9 Oregon 0.591
10 Colorado 0.621
# ... with 11 more rowsmeasles_df %>% filter(state == "Arkansas")
# A tibble: 567 x 2
state mmr_threshold
<fct> <fct>
1 Arkansas Above
2 Arkansas Above
3 Arkansas Below
4 Arkansas Below
5 Arkansas Below
6 Arkansas Below
7 Arkansas Below
8 Arkansas Below
9 Arkansas Below
10 Arkansas Below
# ... with 557 more rows最後にアーカンソー州が異常に少なかったので、何かの間違いかと思いましたが、やっぱり少なかった。。。
グラフで表すとこんな感じです。
measles_df %>% group_by(state) %>% summarise(mmr = mean(mmr_threshold == "Above")) %>% mutate(state = fct_reorder(state, mmr)) %>% ggplot(aes(state, mmr, fill = state)) + geom_col(show.legend = FALSE) + scale_y_continuous(labels = scales::percent_format()) + coord_flip()
3.モデリング
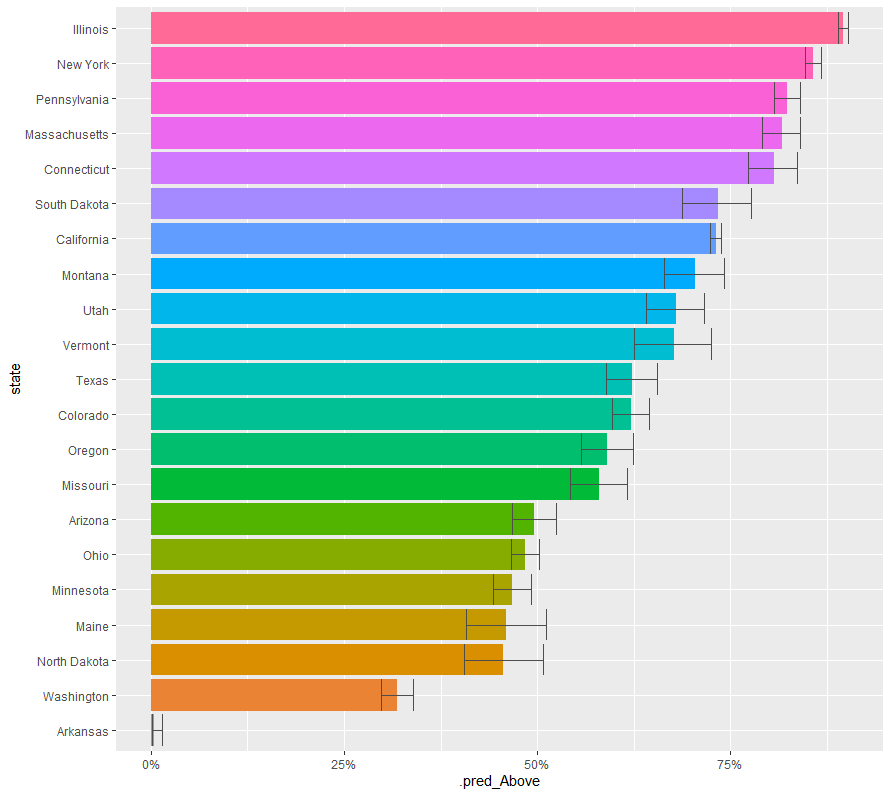
まずは、glmで。
# Getting started with tidymodels
library(tidymodels)
glm_fit <- logistic_reg() %>%
set_engine("glm") %>%
fit(mmr_threshold ~ state, data = measles_df)
tidy(glm_fit) %>% filter(p.value < 0.05)
new_schools <- tibble(state = unique(measles_df$state))
mean_pred <- predict(glm_fit,
new_data = new_schools,
type = "prob")
conf_int <- predict(glm_fit,
new_data = new_schools,
type = "conf_int")
schools_result <- new_schools %>%
bind_cols(mean_pred) %>%
bind_cols(conf_int)
schools_result %>%
mutate(state = fct_reorder(state, .pred_Above)) %>%
ggplot(aes(state, .pred_Above, fill = state)) +
geom_col(show.legend = FALSE) +
geom_errorbar(aes(ymin = .pred_lower_Above,
ymax = .pred_upper_Above),
color = "gray30") +
scale_y_continuous(labels = scales::percent_format()) +
coord_flip()
次にベイジアンモデルも。
# Trying another model Baysian model
library(rstanarm)
options(mc.cores = parallel::detectCores())
prior_dist <- student_t(df = 2)
stan_fit <- logistic_reg() %>%
set_engine("stan",
prior = prior_dist,
prior_intercept = prior_dist) %>%
fit(mmr_threshold ~ state, data = measles_df)
bayes_pred <- predict(stan_fit,
new_data = new_schools,
type = "prob")
bayes_int <- predict(stan_fit,
new_data = new_schools,
type = "conf_int")
bayes_result <- new_schools %>%
bind_cols(bayes_pred) %>%
bind_cols(bayes_int)
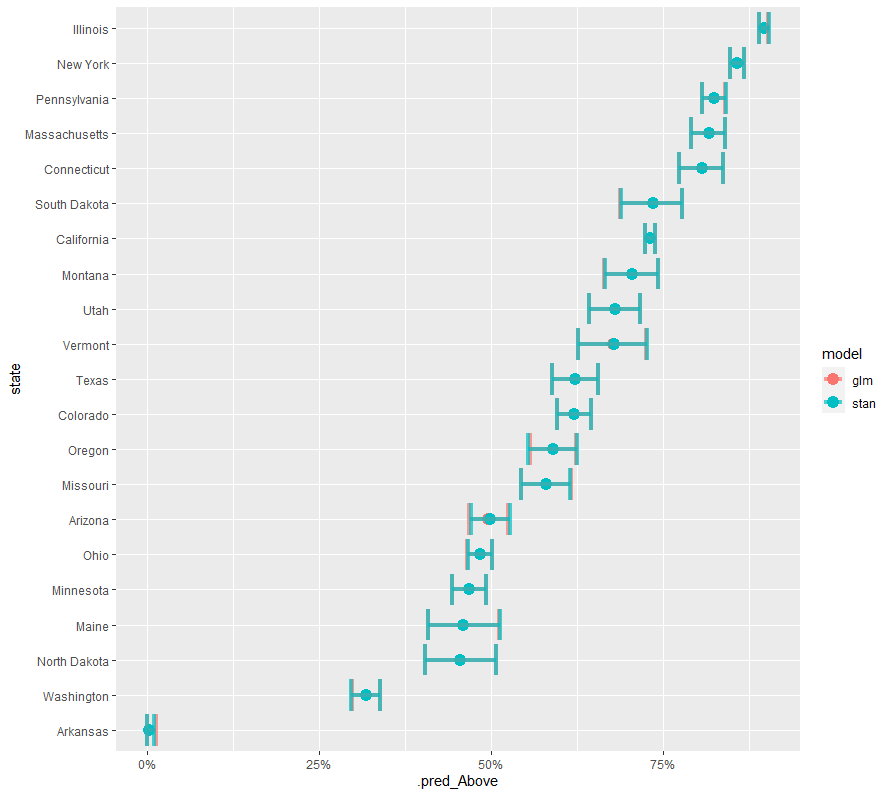
最後に2つのモデルの結果をまとめて表示。glmとベイジアンですごく一致!
schools_result %>%
mutate(model="glm") %>%
bind_rows(bayes_result %>%
mutate(model ="stan")) %>%
mutate(state = fct_reorder(state, .pred_Above)) %>%
ggplot(aes(state, .pred_Above, color = model)) +
geom_point(size = 4) +
geom_errorbar(aes(ymin = .pred_lower_Above,
ymax = .pred_upper_Above),
size = 1.5, alpha = 0.7) +
scale_y_continuous(labels = scales::percent_format()) +
coord_flip()